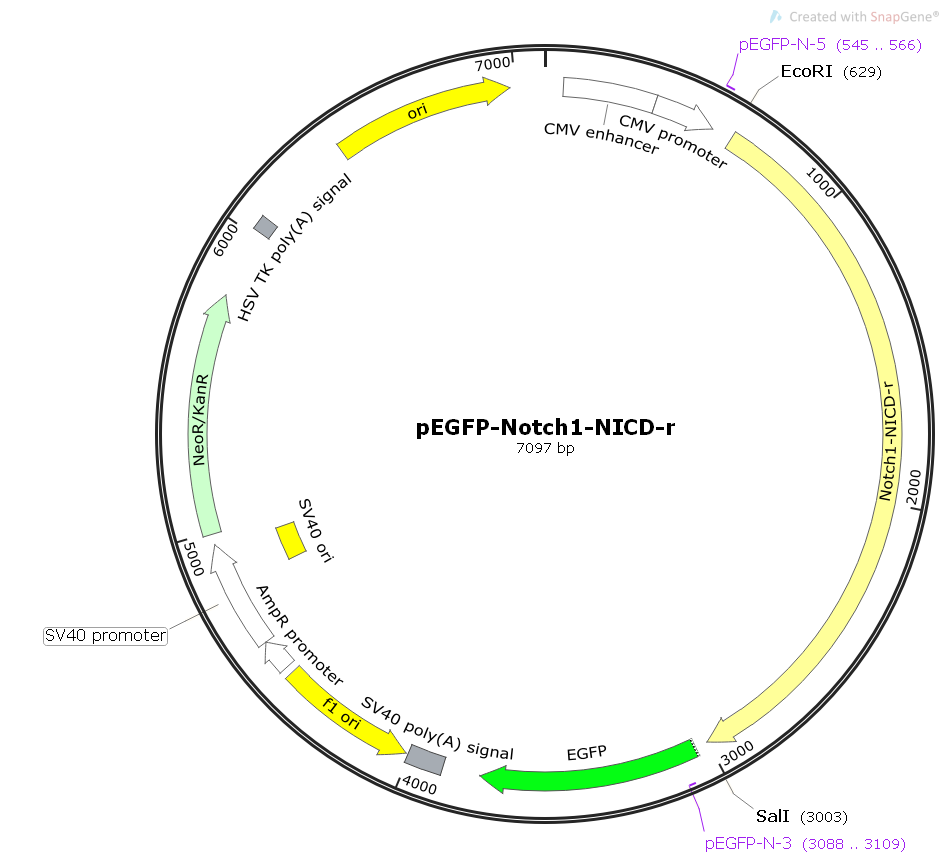
pEGFP-Notch1-NICD-r
Catalog No. PVT10478
Packing 2ug
pEGFP-Notch1-NICD-r Information
Promoter: CMV promoter
Replicator: pUC ori, F1 ori, SV40 ori
Terminator: SV40 poly (A) signal
Plasmid classification: gene library plasmid; rat gene plasmid; rat eukaryotic plasmid.
Plasmid size: 7079bp
Plasmid tagging: C-EGFP
Prokaryotic resistance: Kan (50 g/ml)
Selection marker: Neo/G418
Cloning strains: E. coli DH5 and E.
Culture conditions: 37 C, LB, aerobic.
Expression host: mammalian cells such as 293T
Culture conditions: 5%CO2, 37 C
Induction mode: no need to induce
5'sequencing primers: pEGFP-N-5 (TGGGAGGTCTATATAAGCAGAG)
3'sequencing primers: pEGFP-N-3 (CGTCGCCGTCCAGCTCGACCAG)
pEGFP-Notch1-NICD-r Description
pEGFP-Notch1-NICD-r expresses EGFP-Notch1-NICD fusion protein under the action of CMV strong promoter. The transmembrane macromolecule Notch1 receptor is mainly involved in the selection of cell fate during development. The active fragment-cytoplasmic domain (NICD) of G1743 and V1744 can be transported into the nucleus and activate the expression of target genes. Notch1 plays an important role in cell proliferation, differentiation, programmed death, morphogenesis and organogenesis during development. Cheng. Our constructed pEGFP-Notch1-NICD-r plasmid was transfected into COS-7L cells and green fluorescent protein was observed as nuclear localization, suggesting that the cloned NICD gene is in line with the further study of the function of NICD in post-division neurons.
pEGFP-Notch1-NICD-r Multiple cloning site

pEGFP-Notch1-NICD-r Sequence
LOCUS Exported 7097 bp ds-DNA circular SYN 04-JUL-2017
DEFINITION synthetic circular DNA
ACCESSION .
VERSION .
KEYWORDS pEGFP-Notch1-NICD-r
SOURCE synthetic DNA construct
ORGANISM synthetic DNA construct
REFERENCE 1 (bases 1 to 7097)
AUTHORS yinchu
TITLE Direct Submission
JOURNAL Exported Tuesday, July 4, 2017 from SnapGene Viewer 3.3.4
http://www.snapgene.com
FEATURES Location/Qualifiers
source 1..7097
/organism="synthetic DNA construct"
/mol_type="other DNA"
enhancer 61..364
/label=CMV enhancer
/note="human cytomegalovirus immediate early enhancer"
promoter 365..568
/label=CMV promoter
/note="human cytomegalovirus (CMV) immediate early
promoter"
misc_feature 637..3002
/label=Notch1-NICD-r
/note="Rattus norvegicus notch 1 (Notch1), mRNA
NCBI Reference Sequence: NM_001105721.1
"
CDS 3043..3762
/codon_start=1
/product="enhanced GFP"
/label=EGFP
/note="mammalian codon-optimized"
/translation="MVSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTL
KFICTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDD
GNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIK
VNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLL
EFVTAAGITLGMDELYK"
polyA_signal 3885..4006
/label=SV40 poly(A) signal
/note="SV40 polyadenylation signal"
rep_origin complement(4013..4468)
/direction=LEFT
/label=f1 ori
/note="f1 bacteriophage origin of replication; arrow
indicates direction of (+) strand synthesis"
promoter 4495..4599
/gene="bla"
/label=AmpR promoter
promoter 4601..4958
/label=SV40 promoter
/note="SV40 enhancer and early promoter"
rep_origin 4809..4944
/label=SV40 ori
/note="SV40 origin of replication"
CDS 4993..5787
/codon_start=1
/gene="aph(3')-II (or nptII)"
/product="aminoglycoside phosphotransferase from Tn5"
/label=NeoR/KanR
/note="confers resistance to neomycin, kanamycin, and G418
(Geneticin(R))"
/translation="MIEQDGLHAGSPAAWVERLFGYDWAQQTIGCSDAAVFRLSAQGRP
VLFVKTDLSGALNELQDEAARLSWLATTGVPCAAVLDVVTEAGRDWLLLGEVPGQDLLS
SHLAPAEKVSIMADAMRRLHTLDPATCPFDHQAKHRIERARTRMEAGLVDQDDLDEEHQ
GLAPAELFARLKASMPDGEDLVVTHGDACLPNIMVENGRFSGFIDCGRLGVADRYQDIA
LATRDIAEELGGEWADRFLVLYGIAAPDSQRIAFYRLLDEFF"
polyA_signal 6019..6066
/label=HSV TK poly(A) signal
/note="herpes simplex virus thymidine kinase
polyadenylation signal (Cole and Stacy, 1985)"
rep_origin 6395..6983
/direction=RIGHT
/label=ori
/note="high-copy-number ColE1/pMB1/pBR322/pUC origin of
replication"
ORIGIN
1 tagttattaa tagtaatcaa ttacggggtc attagttcat agcccatata tggagttccg
61 cgttacataa cttacggtaa atggcccgcc tggctgaccg cccaacgacc cccgcccatt
121 gacgtcaata atgacgtatg ttcccatagt aacgccaata gggactttcc attgacgtca
181 atgggtggag tatttacggt aaactgccca cttggcagta catcaagtgt atcatatgcc
241 aagtacgccc cctattgacg tcaatgacgg taaatggccc gcctggcatt atgcccagta
301 catgacctta tgggactttc ctacttggca gtacatctac gtattagtca tcgctattac
361 catggtgatg cggttttggc agtacatcaa tgggcgtgga tagcggtttg actcacgggg
421 atttccaagt ctccacccca ttgacgtcaa tgggagtttg ttttggcacc aaaatcaacg
481 ggactttcca aaatgtcgta acaactccgc cccattgacg caaatgggcg gtaggcgtgt
541 acggtgggag gtctatataa gcagagctgg tttagtgaac cgtcagatcc gctagcgcta
601 ccggactcag atctcgagct caagcttcga attctgatgc tgctgtcccg caagcgcagg
661 cggcagcatg gccagctctg gttccctgag ggtttcaaag tgtcagaggc cagcaagaag
721 aagcggagag aacccctcgg cgaggactca gtcggcctca agcccctgaa gaacgcctca
781 gatggtgccc tgatggacga caatcagaac gagtgggggg acgaagacct ggagaccaag
841 aagttccggt ttgaggaacc agtggttctc cctgacctgg atgatcagac tgaccaccgg
901 cagtggaccc agcagcacct ggatgccgct gacctacgtg tgtctgccat ggccccaacg
961 ccgcctcagg gggaggtaga tgctgactgc atggacgtca atgttcgagg accagatggc
1021 ttcactcccc taatgattgc ctcctgcagc ggagggggcc tggagacagg caacagtgag
1081 gaagaagaag atgcacctgc tgtcatctcc gacttcatct atcagggtgc cagcttgcac
1141 aaccagacgg accgcacagg ggagactgcc ctgcacctgg ctgcccgata ctctcgttca
1201 gatgctgcca agcgcttgct ggaggccagc gcagatgcca acatccaaga caacatgggg
1261 cgtaccccat tacatgccgc tgtttctgca gacgctcagg gtgtcttcca gatcctgctc
1321 cggaacagag ccacagatct ggatgcccga atgcatgatg gcacaacccc tctgatcctg
1381 gcggcacgcc tggccgtgga aggcatgcta gaggacctca tcaactctca cgctgatgtc
1441 aatgctgtgg atgacctagg caagtcagct ctgcactggg cagccgctgt gaacaatgtg
1501 gacgctgctg ttgtgctcct gaagaacgga gccaacaaag acatgcagaa caacaaggag